Forest for the Trees
A Python Tree Structure Survey
or what I learned over the weekend
Trees the Basics
If I had a real use for trees, I'd use the ETE2 library.
Want to pass that interview test question, how about:
from ete2 import Tree
t = Tree("(A,(B,C));")
Or this little snippet seems to get a lot of play:
from collections import defaultdict
def tree():
return defaultdict(tree)
Though, honestly, I don't really appreciate the above; and if we're just trying to get through an interview, this following is probably a much better answer:
tree = [0,[1,[2,3]]]
Of course, you might want to touch base with someone who's actually been interviewed (or better yet, conducts interviews for a living) to get a better idea of what to say during an actual interview.
So, perhaps I should say, if I was going to work on a project, in which I thought I needed a tree class, I'd read up on ete2 and probably end up using the Newick Tree format to track my data.
Newick Trees
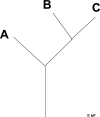
This is a simple tree.
The same data in Newick format is:
(A,(B,C);
So, () for a node, and X wherever data occurs.
Probably not overly clear, but the search term you're looking for is Newick Trees. They even have viewers for these things, so it's a well established standard and a person couldn't go wrong utilizing them in a project via ete2.
I also looked at networkx briefly, but trees are sort of tagged on as an afterthought in that library (once again, in my so ever humble opinion, and as much as
I like networkx for cool graphs), whereas ete2 has commands for everything I could ever possibly want out of a tree library; such as:
swap_children()
traverse()
get_anscestors()
get_children()
prune()
remove_child()
remove_sisters()
And so on and so forth.
Also, the graphing interface looks pretty, kick, assuming hip folks on the Internet still use such terms. Want to graph a DNA sequence? Well, if so, ete2 is probably the library you're looking for. Well, that or biopython. But I haven't really spent much time (or any time for that matter) with biopython, so I really couldn't say.
Anyhow, enough for the intro.
List Processing
I wanted to look into trees, because folks kept on mentioning them in the articles I was reading, but I'd never had cause to use them and I didn't know if that was because I was ignorant of their nature.. or I was ignorant of them because their uses were trivial.
Anyway, after having looked at them for a second or two (I'm a great one for jumping to conclusions), their structure sort of reminded me of Lisp and/or Haskell: two languages I have some ability to read (if the keywords are small and simple enough), but which I won't claim any expertise. (I view using monads as a akin to multiplying two numbers by first converting them to logs, adding said numbers together, and then converting the result back through exponential multiplication. So, clearly, my ignorance is legion.)
Thus it was with little difficulty that I saw the relationship between:
Lisp: (+ 1 1)
Newick: (A,1,1)
Python: [operator.add,1,1]
Or in other words:
Lisp: (+ 1 (+ 1 1) ~= Python: [operator.add,1,[operator.add,1,1]]
Or close enough for me. (I was at a party the other night and one my fellow guests said something along the lines of "Twice three, so seven." I knew exactly what they meant. Or in other words, close enough for me. Unlike my compiler, I accept all sorts or malformed syntax.)
Anyway, what was I saying?
Oh, right, the two are close enough for me, so to process the operator list shown above, I went with:
def parse_list(a):
'''given that a is of the recursive form [operator, int, int]
with any int being replaceable by the whole
then, this bad boy returns the integer reduction of a
'''
a = copy.deepcopy(a)
for n in [0,1,2]:
if isinstance(a[n], list):
a[n] = parse_list(a[n])
r = a[0](a[1],a[2])
return r
Without the copy.deepcopy(), this throws off all sorts of hard to track down bugs that could easily eat up an afternoon of your life, just saying.
And here I need a witty transition, so just imagine that I'm a morbidly depressed robot, as you read the next line, "Life. Don't talk to me about life."
Genetic Programming
I don't know enough about this topic to know if that's the right heading. Genetic Programming, Machine Learning, Random Walk, it's all the same to me.
Essentially, having put together the above parser, I wanted to take the proof of concept a little further. As such, the following function creates an ever more (or less) complicated iterative list that when put through the parser will spit out the sought number (i.e. the goal).
import operator
import random
import copy
def genetic_this_or_that(seed=[operator.add,1,1],
goal=1018
):
'''attempts to find an iterative list of form discussed
that when parsed equals the goal
'''
currentBest = copy.deepcopy(seed)
cB = parse_list(currentBest)
if cB == goal:
return currentBest
nextTry = random_change(copy.deepcopy(seed))
nT = parse_list(nextTry)
if abs(goal - cB) <= abs(goal - nT):
print "RETURN currentBest: %s" % currentBest
return genetic_this_or_that(currentBest,goal)
else:
print "RETURN nextTry: %s" % nextTry
return genetic_this_or_that(nextTry,goal)
goal=10
#goal=1018 #which yields an iterative overflow about half the time
solutionSet = genetic_this_or_that(goal=goal)
print "\n\n%d Reached:\n%s" % (goal, solutionSet)
When solving for 1018, the above exceeds Python's (or at least, my setup's) default iteration limit of 1000 about half the time.
But perhaps more importantly, the above requires:
def random_change(inList):
'''makes a random change to a list tree
'''
return random.choice([random_node_to_leaf(inList, random.randint(0,99)),
random_leaf_to_node(inList, simple_random_node())])
Which in turn requires a few more functions, which since I tire of writing, I shall assume are self explanatory.
def simple_random_node():
'''returns a simple random node of form [operator, int, int]
'''
return [random.choice([operator.add, operator.sub, operator.mul]),
random.randint(0,9),
random.randint(0,9)]
def replace_node_with_leaf(aL,newLeaf,node):
'''replaces a node with a leaf
'''
for n in [0,1,2]:
if callable(aL[n]):
if node == 0:
node -= 1
return newLeaf, node #aL[node] = newNode
else:
node -= 1
elif isinstance(aL[n], list):
aL[n],node = replace_node_with_leaf(aL[n],newLeaf,node)
return aL,node
And since I also tire of refactoring what is essentially throwaway code for presentation, I do believe I shall stop the cut and paste there.
Suffice to say, I found it easier to replace a leaf with a node than a node with a leaf, so (in my opinion), the more complicated code is shown.
Anyway, hope you found the above informative... or if not informative, at least interesting... or if not interesting, at least not too inane.
And with that said, I will say adieu.
Or why not say it?
I'm currently looking for a job.
After 25 years, I've finally retired.
So, Hire me... or don't.
I'll be at my keyboard either way tomorrow: the only real difference being whether I work for love or money... so, you know, why not both?
Brett Code
www.paufler.net
#&169; Copyright 2015 Brett Paufler
paufler.net@gmail.com
Terms of Service
